My MSc Thesis
My thesis was focussed on finding the best way to detect invasive fish in lakes using environmental DNA. This is a relatively new technology that can potentially detect hundreds of different organisms that are living in a lake from just one litre of water! My ultimate goal is to work in the ecology or environmental consultancy space - specifically in the terrestrial, freshwater or marine space.
I am currently a biosecurity advocate for the Waikato Regional Council (WRC) and the Ministry for Primary Industries (MPI) where I promote the CHECK-CLEAN-DRY program to community groups, sporting organisations and at major water-sports events. The aim of this program is to educate the public about biosecurity and to ensure that all water users are traveling with clean gear between waterways every time.
I love all things to do with water, I am a fully qualified PADI Dive Master and Rescue diver and I try to get out in the ocean for a surf whenever I can!
Abstract
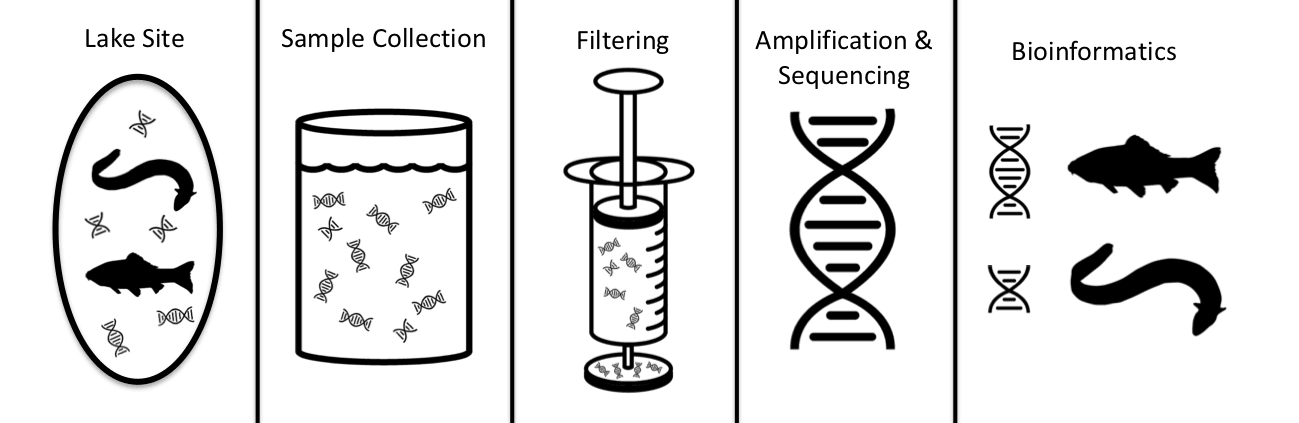
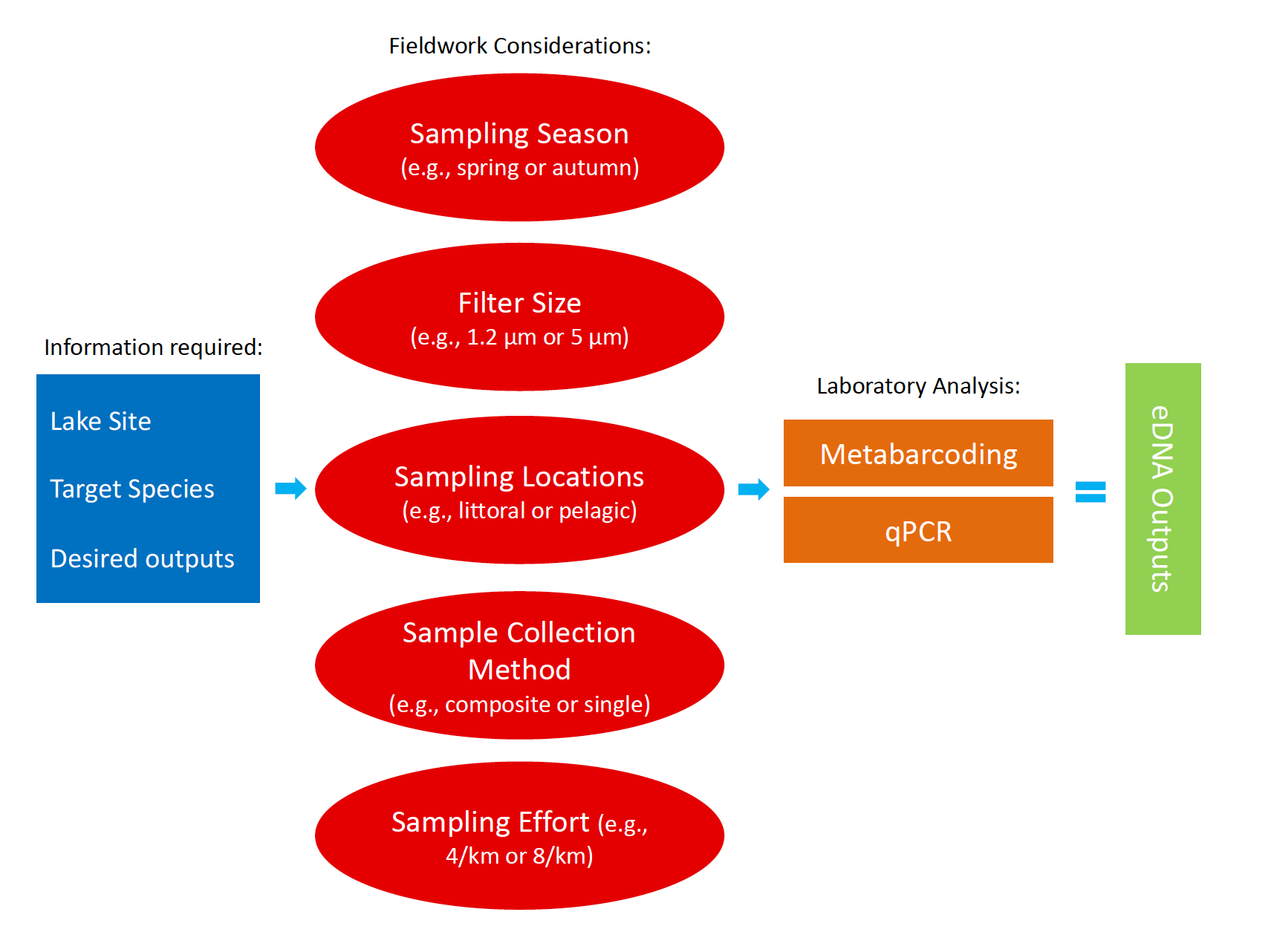
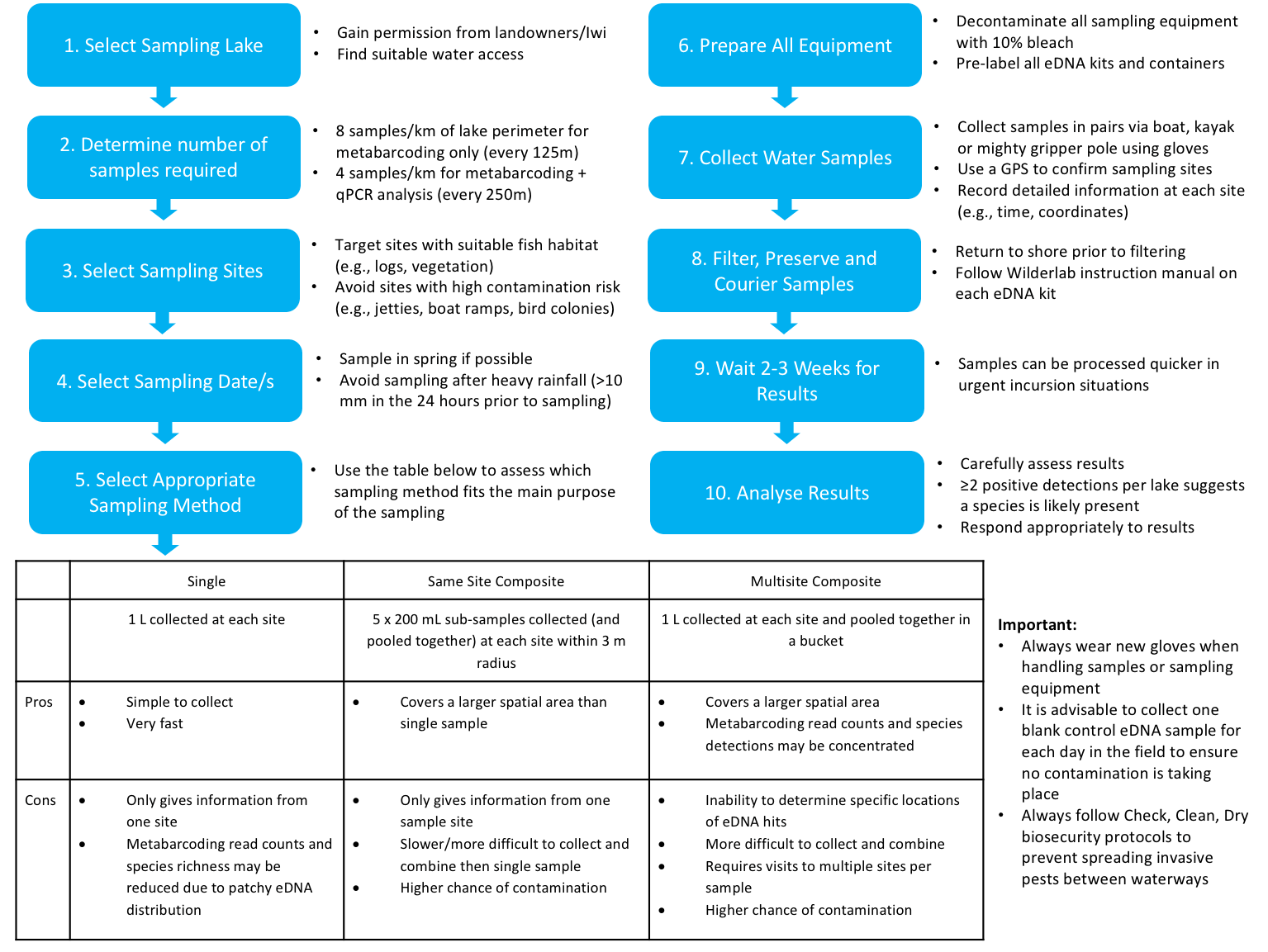
Environmental DNA (eDNA) is a quick, effective and sensitive biomonitoring tool that can be used to detect invasive species at low densities in aquatic habitats. The aim of my study was to develop a standard operating protocol (SOP) for the detection of invasive fish in New Zealand’s lowland lakes, specifically koi carp (Cyprinus rubrofuscus). Four separate eDNA surveys between October 2022 and April 2024 sampled 18 lakes in total across the Auckland and Waikato Regions in the North Island of New Zealand. I evaluated different aspects of eDNA sampling including sample collection methods, filter sizes, seasonality, laboratory analysis methods, along with the optimal number of samples and spatial locations to filter water from. My study found that eDNA was a simple, reliable, and effective tool that can be easily used to detect koi and other pest species at low densities across a wide variety of lakes. The efficacy of eDNA sampling was proven during a real-world koi incursion at Lake Waiwhakareke, where I successfully detected koi for the first time in this lake at very low densities using eDNA. Overall, I found that a coarser (5 μm) filter size outperformed the finer (1.2 μm) filter in every comparison and that the qPCR laboratory analysis method was slightly superior to the metabarcoding method, although it has the disadvantage of only being able to detect a single species. My study showed that the multisite composite sample collection method performed marginally better than the single collection overall, across 13 lakes in the Auckland Region. Targeting the littoral margins of the lakes was found to be significantly better in terms of metabarcoding read counts and species detections than sampling the pelagic/mid-lake regions at both Lake Puketirini and Lake Waiwhakareke. In the spatio-temporal study in the Waikato, I found that there was a higher degree of variation between the two seasons sampled (Spring and Autumn) than there was between the three lakes. The spring season performed best in terms of fish detections and metabarcoding read counts. In terms of the sampling number and density, I found that eight samples per kilometre of lake perimeter was found to be optimal to detect koi carp at very low densities at Lake Waiwhakareke and Lake Puketirini. However, further research is needed to better understand the uncertainties involved with eDNA sampling, including the amount of sampling required to provide high levels of confidence in the results. Using negative and positive controls at the time of sampling and during laboratories analyses, in addition to species distribution modelling may help ascertain the sensitivities of eDNA detection with a greater level of certainty.



Thesis Supporters
A massive thank you to everyone below who made my thesis a reality!
My thesis supervisor, Frank Burdon
The Department of Conservation, the primary financial supporter, who provided me with financial support, technical advice, eDNA kits, equipment and support in the field. Particularly Kerry Bodmin and Nigel Binks.
The Ministry for Primary Industries, who provided eDNA kits and financial support for the Lake Puketirini study.
High Performance Sport NZ for awarding me the Prime Ministers Scholarship.
Freemasons New Zealand for awarding me the Post-Graduate Scholarship.
The University of Waikato for providing me with a Masters Scholarship.
Sargood Bequest for providing me with eDNA kits.
The Mazda Foundation for providing eDNA kits.
The Hillary Jolly Scholarship committee for awarding me the Hillary Jolly Scholarship.
The New Zealand Biosecurity Institute for awarding me their Biosecurity Scholarship.
Forest and Bird for awarding me the Valder Grant to purchase eDNA kits.
The landowners who gave me permission to access their lakes.
Dr Shaun Wilkinson and his team at Wilderlab providing me with advice and laboratory analysis.
Ange Chaffe and her team at Auckland Council for generously providing me the data from the 13 Auckland Region Lakes.
Adam Daniels from Fish and Game, who provided a boat for me to use at Lake Puketirini to collect eDNA samples.
Everyone who came out to help with the sampling.
And most importantly my Mum, Dad and sister, who have fully supported my studies alongside my sporting endeavours through the highest highs and lowest lows. Your collective support has allowed me to pursue my passion for conservation, biosecurity and aquatic environments and allowed my research dreams to become realities, I would not have been able to attempt this thesis alongside my sporting aspirations without your emotional and financial support. Thank you.
Full Thesis
Currently the full thesis is under embargo until January 2027, but I will be publicly publishing sections of my thesis in mid-late 2025.



